Example 1: Multiple Sample KEGG View
This example shows the multiple sample/state integration with Pathview KEGG vieww. Data files are pre loaded and the options have been preset as below. Data files used in this example are Gene Data and Compound Data. In this example Gene Data has 3 samples and Compound Data has 2 samples.
API Query
./pathviewapi.sh --gene_data gse16873.d3.txt --cpd_data sim.cpd.data2.csv --species hsa --gene_id ENTREZ --cpd_id KEGG --pathway_id 00640 --suffix multistatekegg
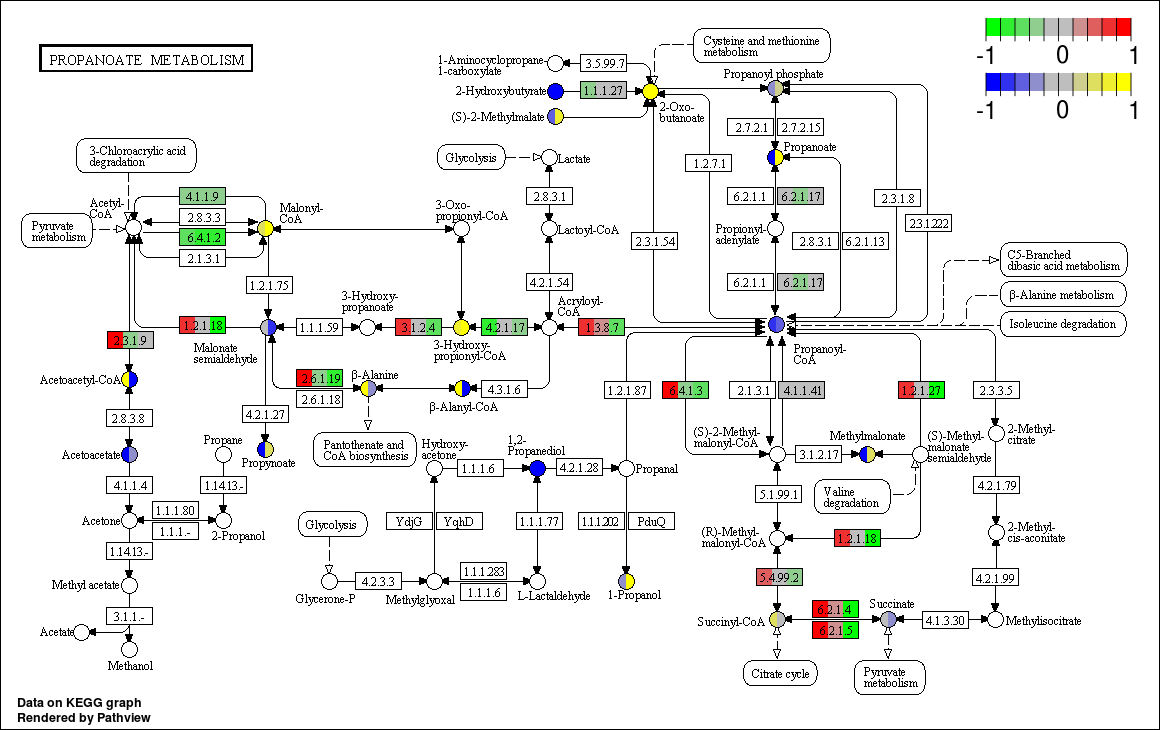
Example 2: Multiple Sample Graphviz View
This example shows the multiple sample/state integration with Pathview Graphviz viewo. Data files are pre loaded and the options have been preset as below. Data files used in this example are Gene Data and Compound Data. In this example Gene Data has 3 samples and Compound Data has 2 samples. Difference on this example is you are generating a Graphviz view by unchecking the kegg view selection box.
API Query
./pathviewapi.sh --gene_data gse16873.3.txt --cpd_data sim.cpd.data1.csv --species hsa --pathway_id 00640 --suffix multi --kegg F --limit_gene -1,2 --cpd_reference 1,2 --cpd_sample 3,4 --gene_reference 1,3,5 --gene_sample 2,4,6
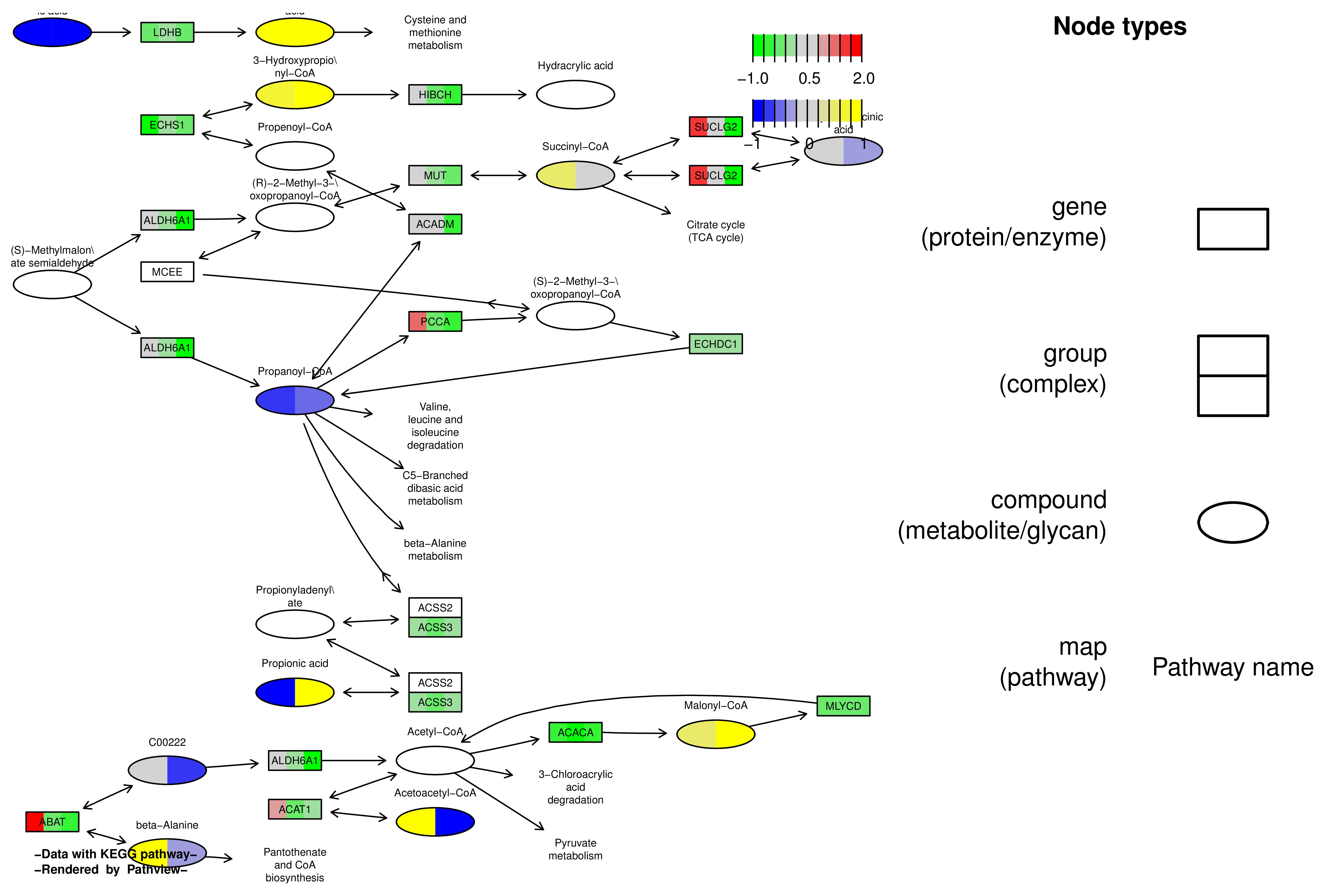
-
GUI Arguments
-
Pathway ID:00640-Propanoate metabolism
-
Species:hsa
-
Kegg Native:FALSE
-
Same Layer:TRUE
-
Multi State:TRUE
-
Match Data:FALSE
-
Gene Limit:-1(min),2(max)
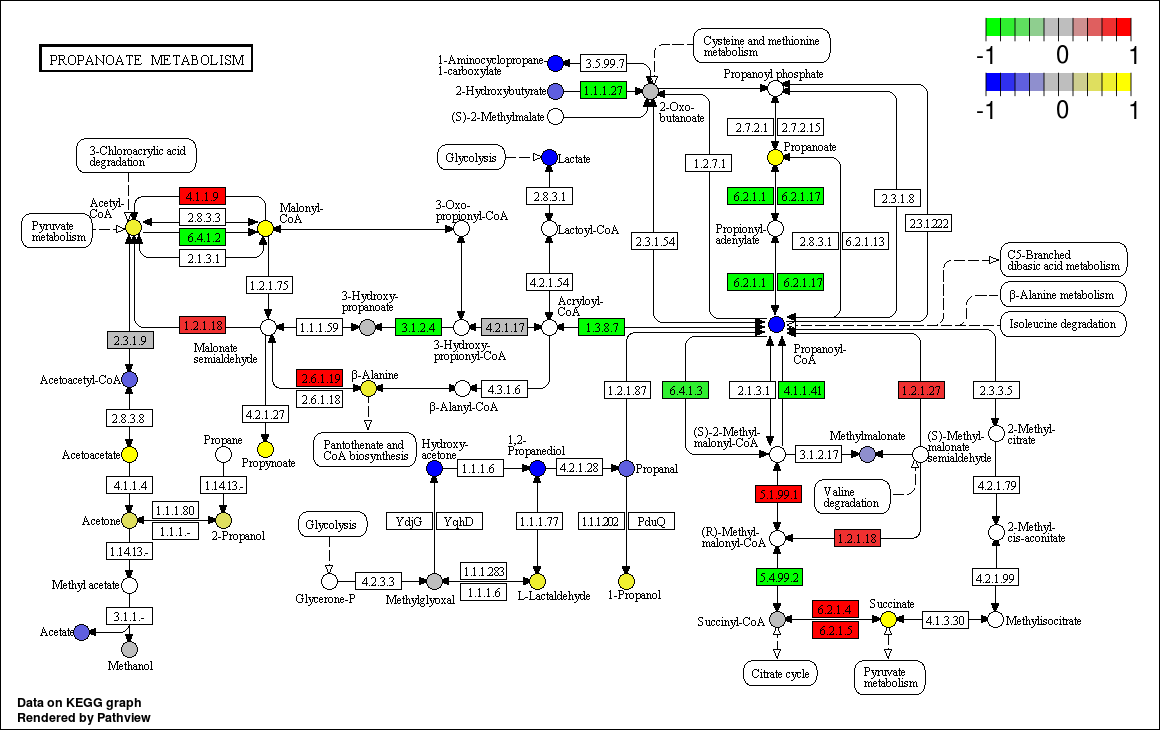
Example 3: ID Mapping
Here's an example showing the ID mapping capability of pathview. Data files are pre loaded and the options have been preset as below (including Gene ID type and Compound ID type). Data files used in this example are Gene Data and Compound Data.
API Query
./pathviewapi.sh --gene_data gene.ensprot.txt --cpd_data cpd.cas.csv --species hsa --gene_id ENSEMBLPROT --cpd_id 'CAS Registry Number' --pathway_id 00640 --suffix IDMapping --limit_gene 3 --limit_cpd 3 --bins_gene 6 --bins_cpd 6
-
GUI Arguments
-
Pathway ID:00640-Propanoate metabolism
-
Species:hsa
-
Gene ID:ENSEMBLPROT
-
Compound ID:CAS Registry Number
-
Kegg Native:TRUE
-
Same Layer:TRUE
-
Limit:Gene:3; Compound: 3
-
Bins:Gene:6; Compound: 6
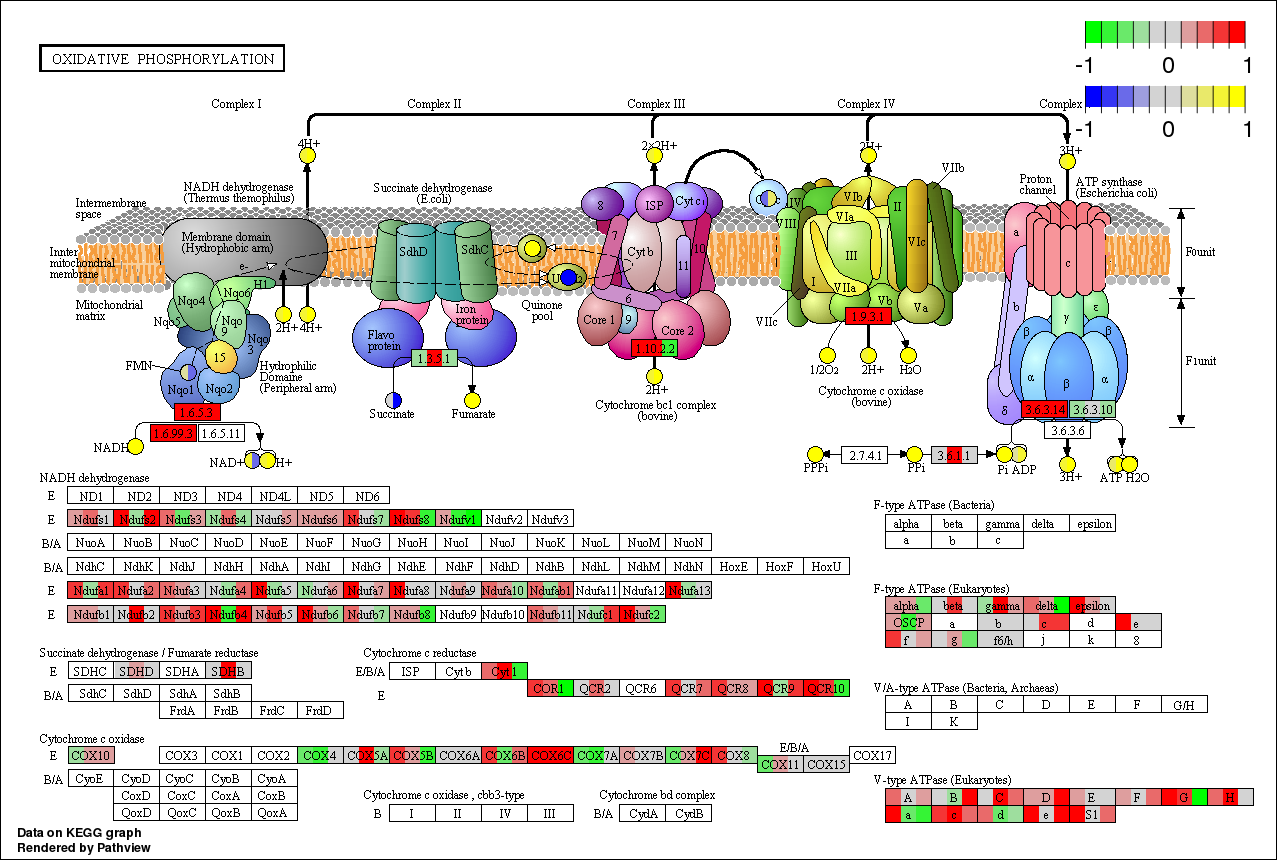
Example 4: Integrated Pathway Analysis (takes longer time)
This example covers a comprehensive workflow including pathway analysis, data integration and visualization. The user options of this analysis are similar to other examples, except that Pathway Selection option is set to Auto instead of Manual. In this case, pathway analysis is done as to determine the target pathways for downstream visualization and interpretation. Since both gene and compound data (inputs) are included, this example goes beyond in that in integrates the two separate regular pathway analysis for gene and compound (intermediate results) into a more powerful meta-analysis (final results). Data files are pre loaded and the options have been preset as below. Data files used in this example are Gene Data and Compound Data.
API Query
./pathviewapi.sh --gene_data gse16873.3.txt --gene_reference 1,3,5 --gene_sample 2,4,6 --cpd_data sim.cpd.gse16873.csv --cpd_reference 1,2 --cpd_sample 3,4 --species hsa --auto_sel T
-
GUI Arguments
-
Species:hsa
-
Kegg Native:TRUE
-
Pathway Selection:Auto
-
Pathway ID:(Not Applicable)