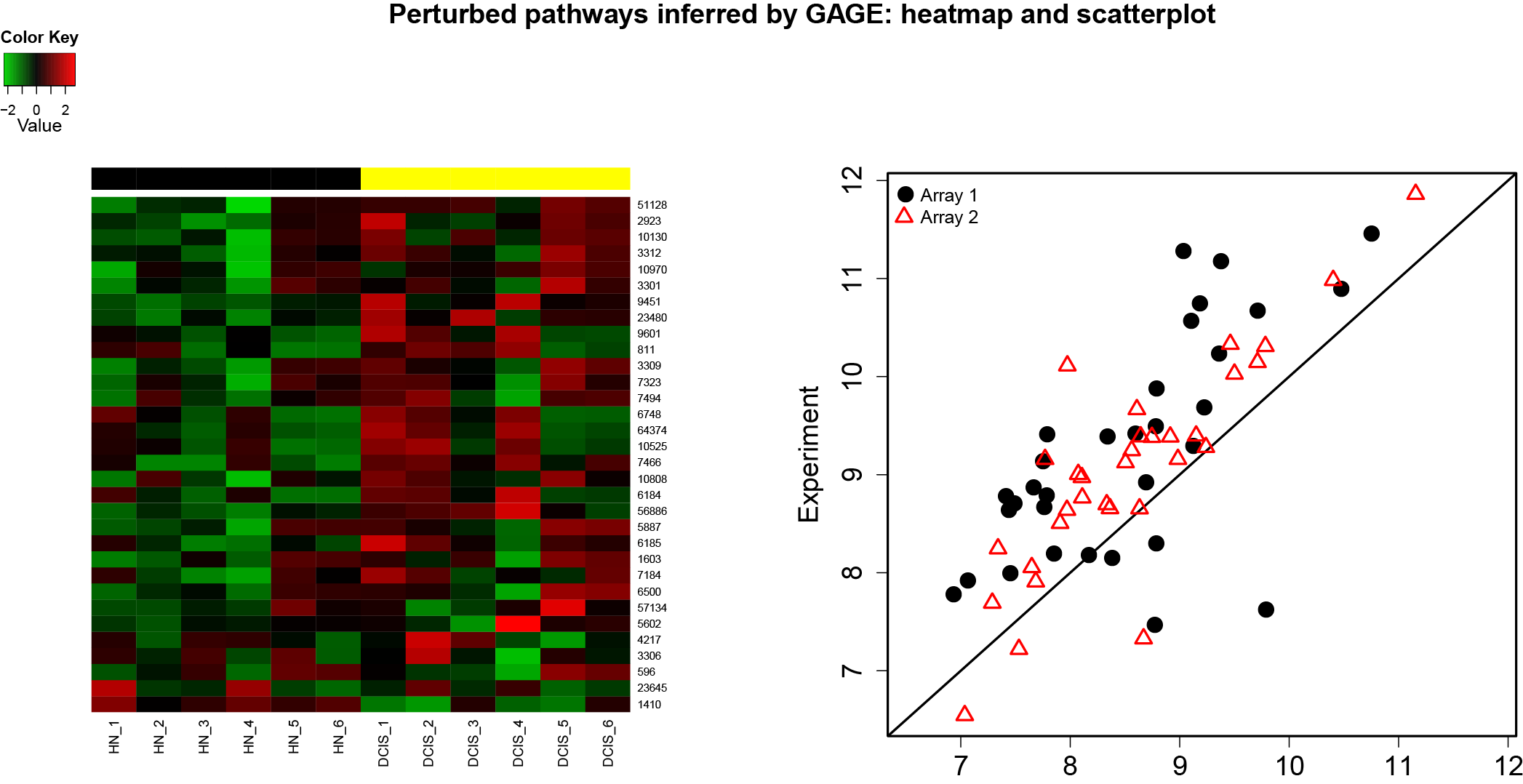
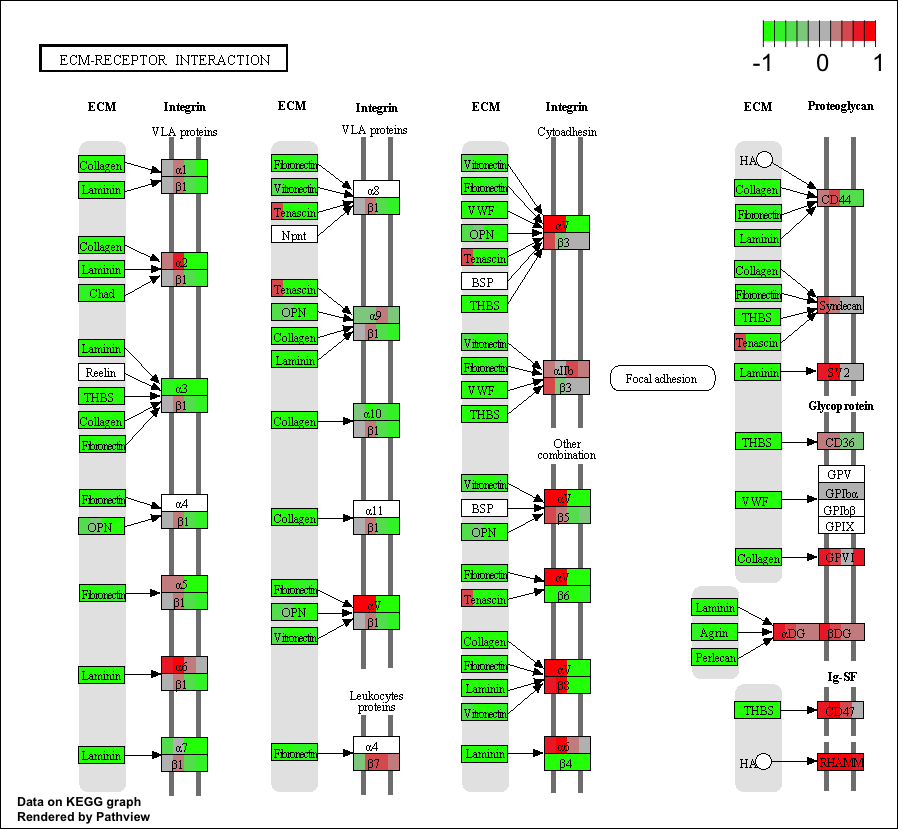
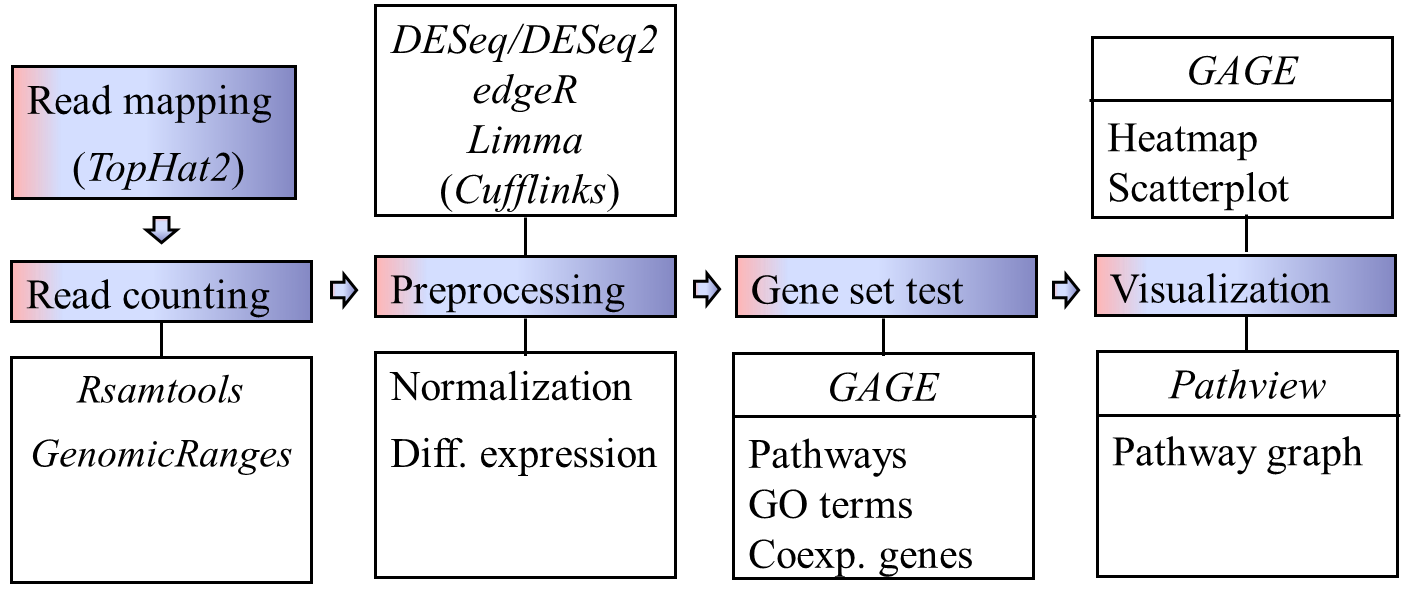
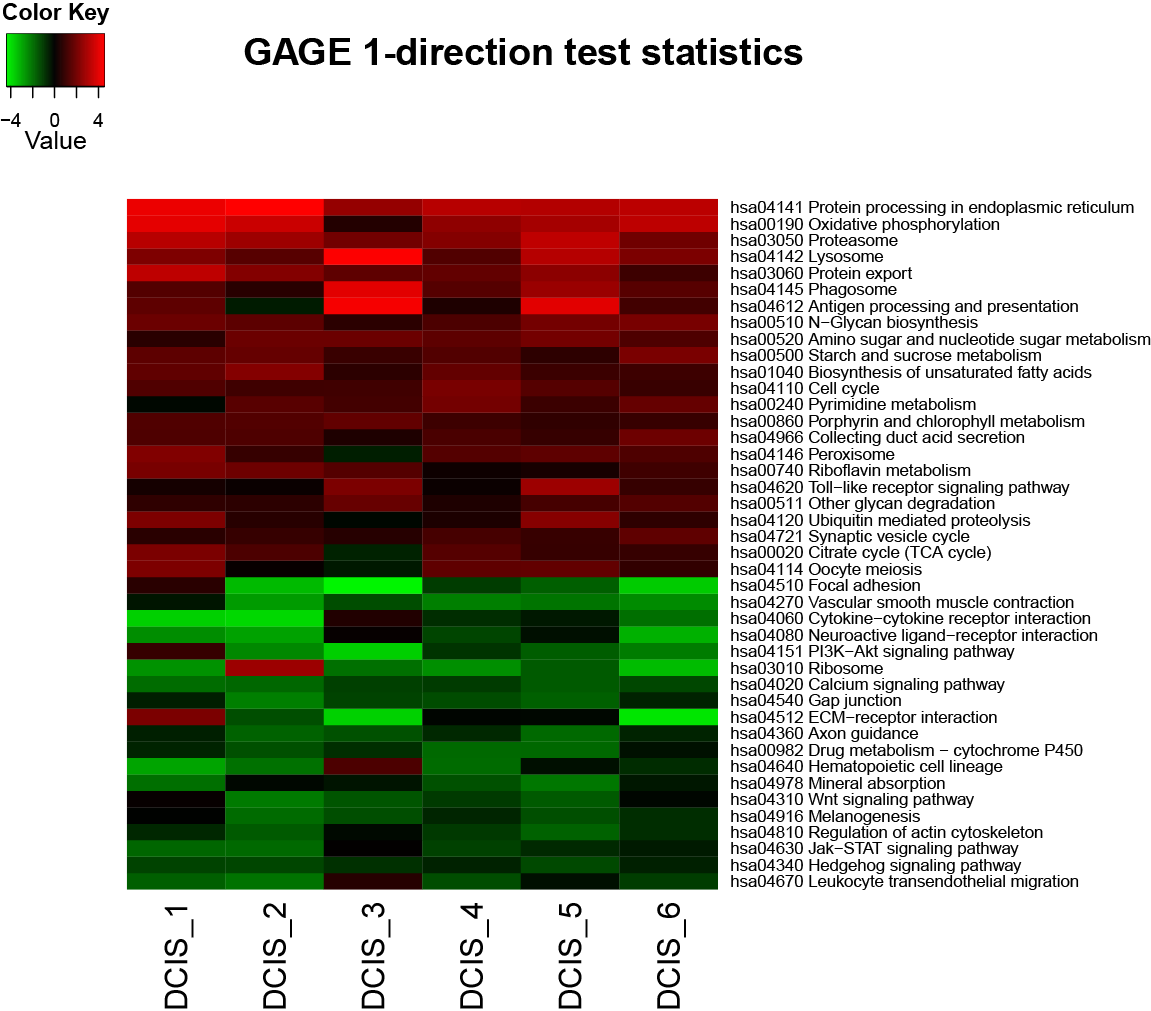
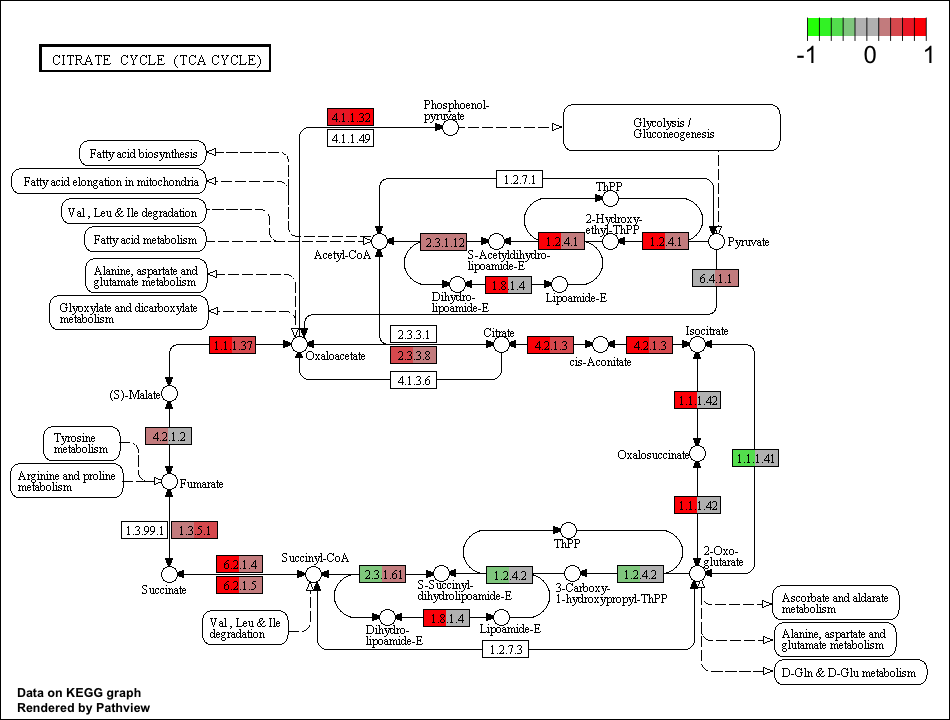
GAGE is an established method for gene set (enrichment or GSEA) or pathway analysis. GAGE is generally applicable to different omics data with robust performance.
GAGE Web provides easy interactive access to GAGE analysis. It features:
- A complete pathway analysis workflow based on GAGE and Pathview
- Support to >3000 species, dozens of molecular IDs, various omics data and gene-set data (KEGG pathways, Gene Ontology, SMPDB etc);
- Over-representation test on preselected gene or molecule lists.
Citations:
Luo W, Friedman M, etc. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics, 2009, 10, pp. 161, doi: 10.1186/1471-2105-10-161
Luo W, Brouwer C. Pathview: an R/Biocondutor package for pathway-based data integration and visualization. Bioinformatics, 2013, 29(14):1830-1831, doi: 10.1093/bioinformatics/btt285
Please cite the GAGE paper and pathview.uncc.edu if you use this website. In addition, please cite the Pathview paper if you the Pathview visualization here. This will help the GAGE and Pathview projects in return.
Contact:
Email us: pathomics@gmail.com