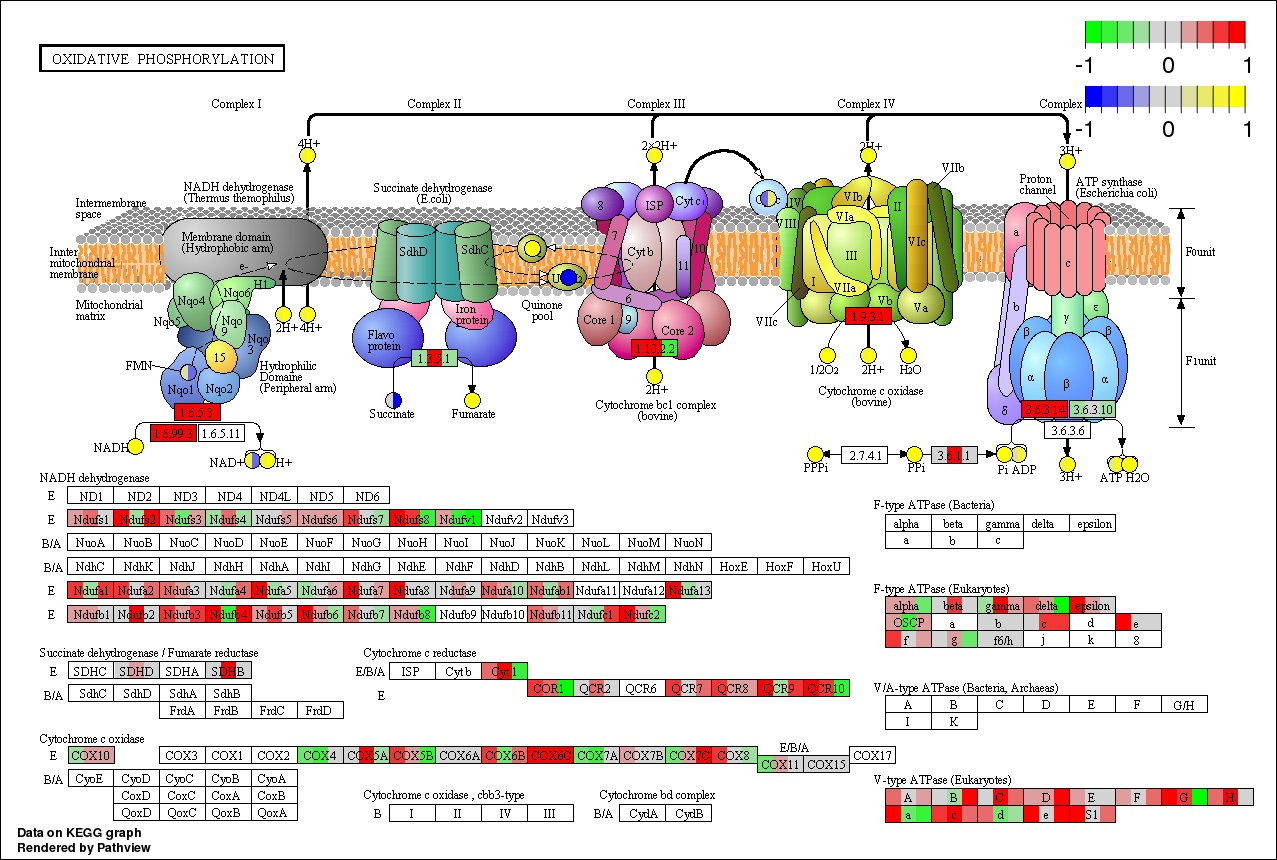
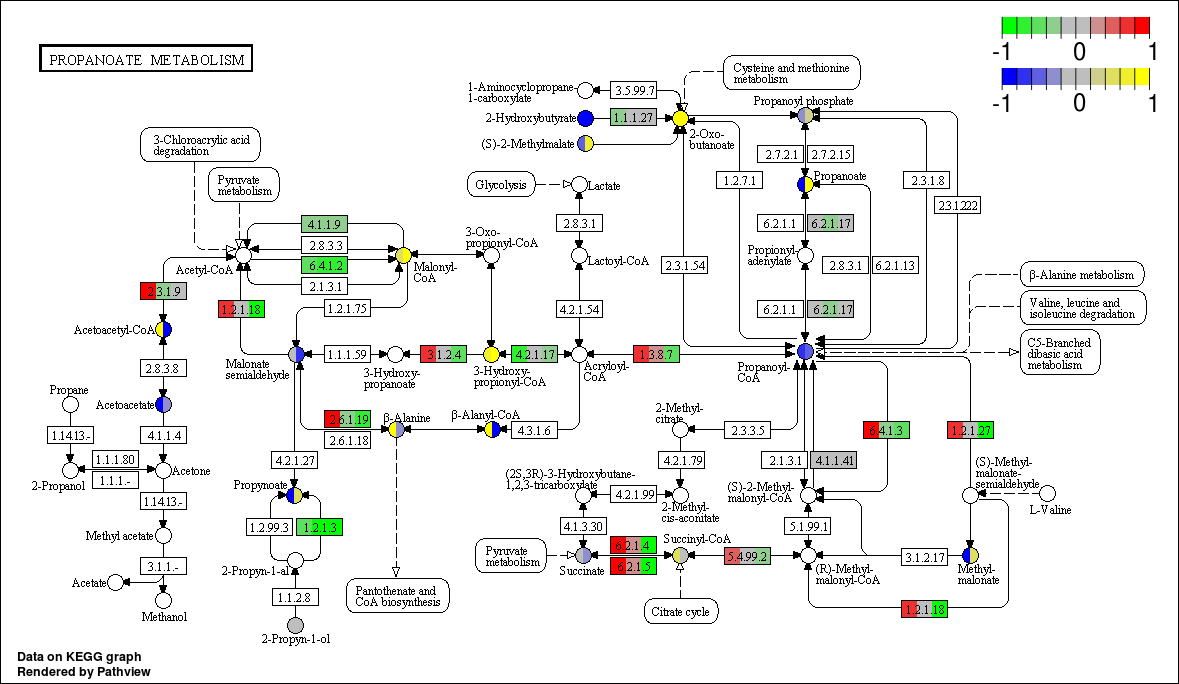
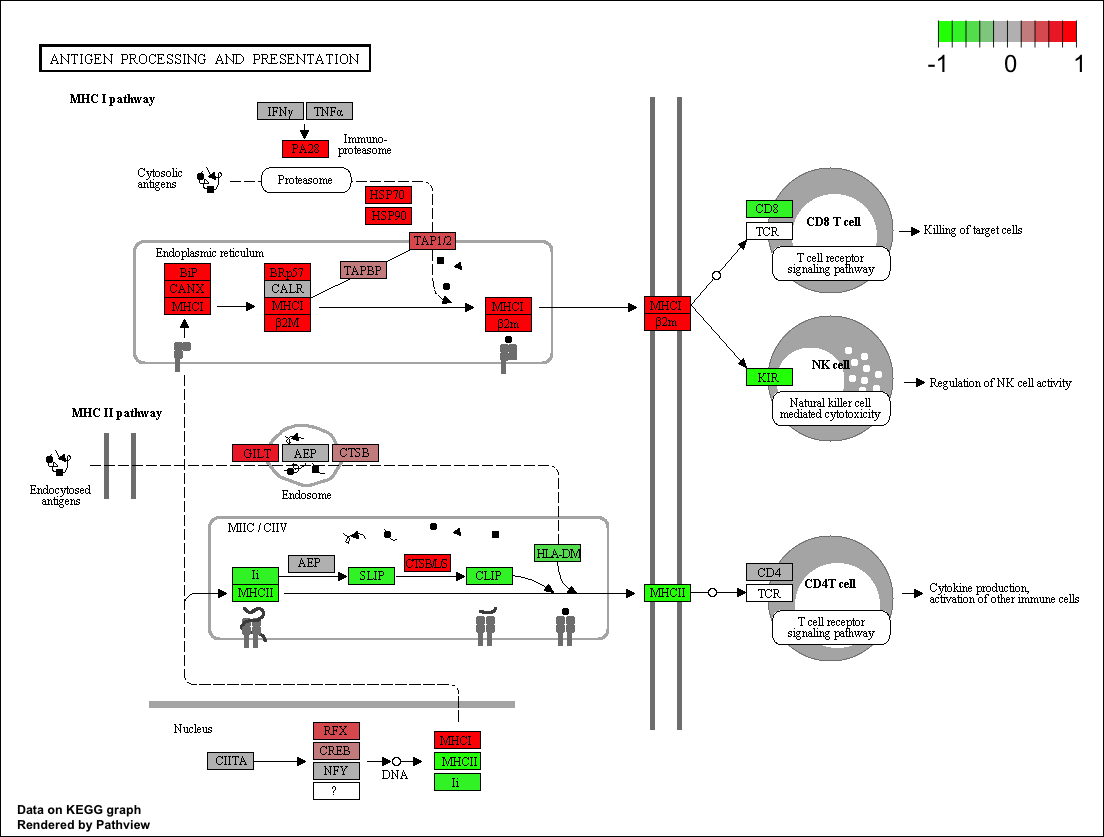
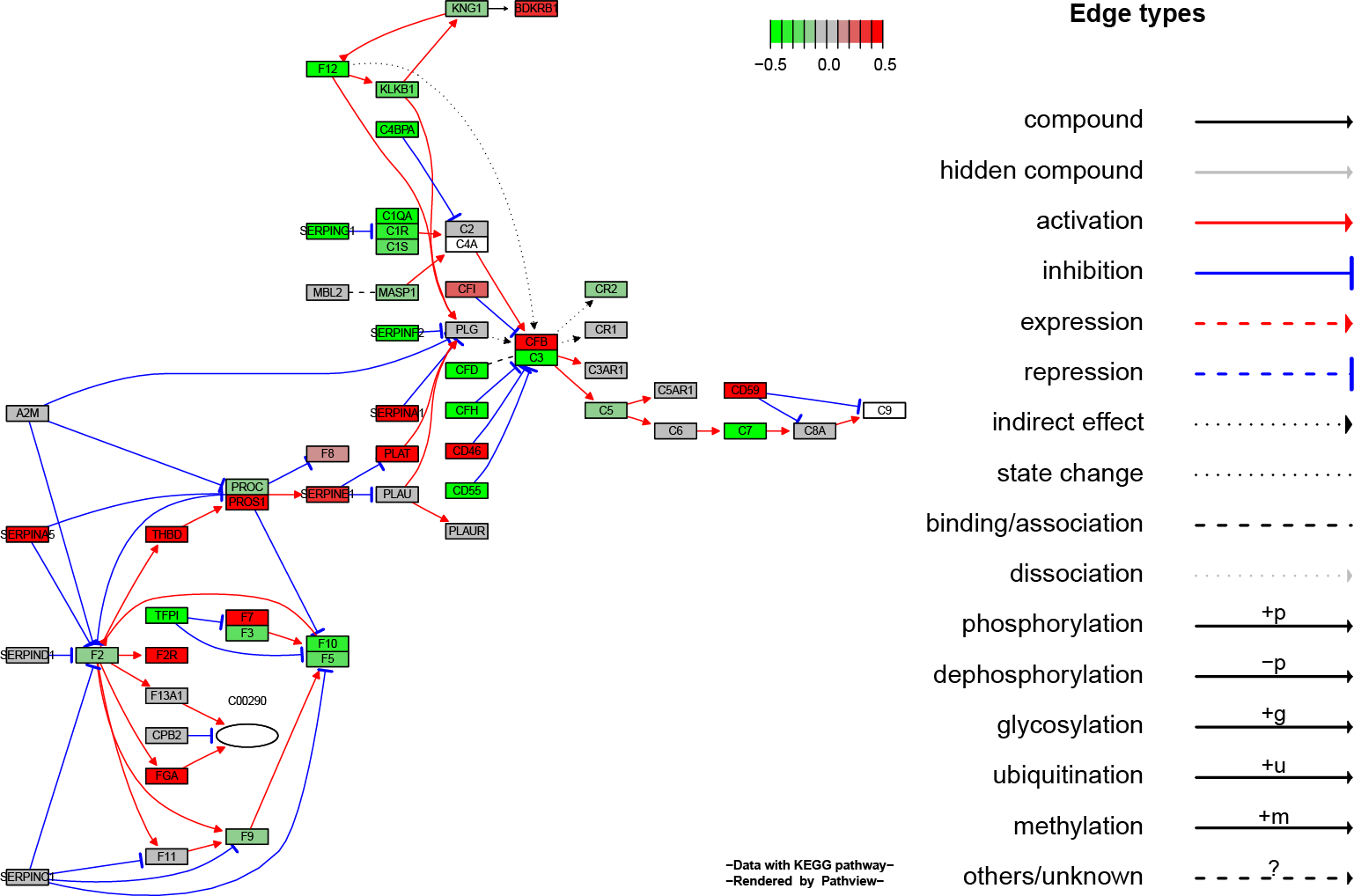
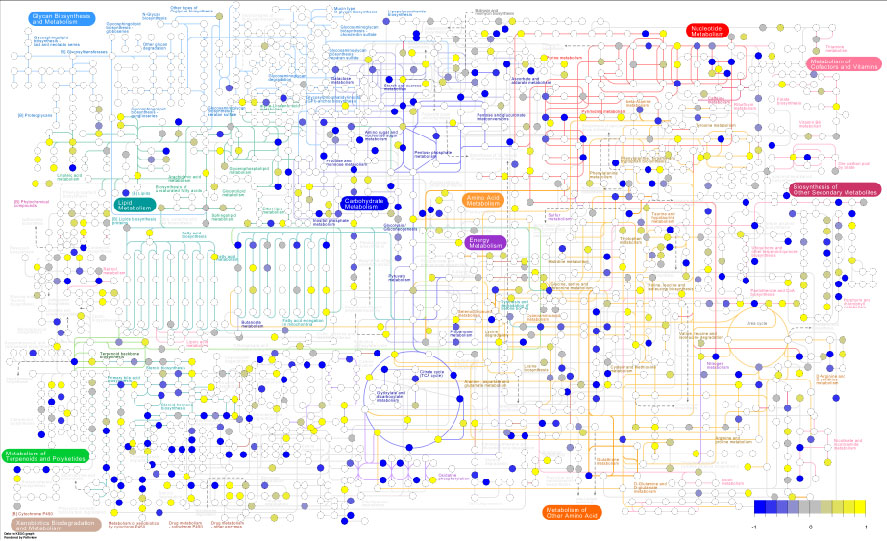
Pathview maps, integrates and renders a wide variety of biological data on relevant pathway graphs.
Citations:
Please cite our paper if you use this website. This will help the Pathview project in return.
Please also cite GAGE paper if you are doing pathway analysis besides visualization, i.e. Pathway Selection set to Auto on the New Analysis page.
Contact:
Email us: pathomics@gmail.com